Professor Kim Seung-il's team succeeded in deciphering the genome of the Korean pine tree Jeongipumsong

- Published in Nature Genetics (IF=31.7), a world-renowned academic journal
- Deciphering the pine standard genome by parent-derived haploid and providing new insights into the evolutionary process of the giant genome and the genetic differences between haploid bodies
A research team led by Professor Kim Seung-il of the Department of Environmental Horticulture in University of Seoul has established a standard dielectric of pine trees reflecting haploid genotype information for the first time in the world, and announced the results of an in-depth study on the evolution process of giant genomes and genetic differences between haploid bodies. Professor Kim Seung-il was the corresponding author, Dr. Jang Min-jeong and Dr. Cho Hye-jeong participated as the first author, and Dr. Park Eung-joon participated as the co-corresponding author.
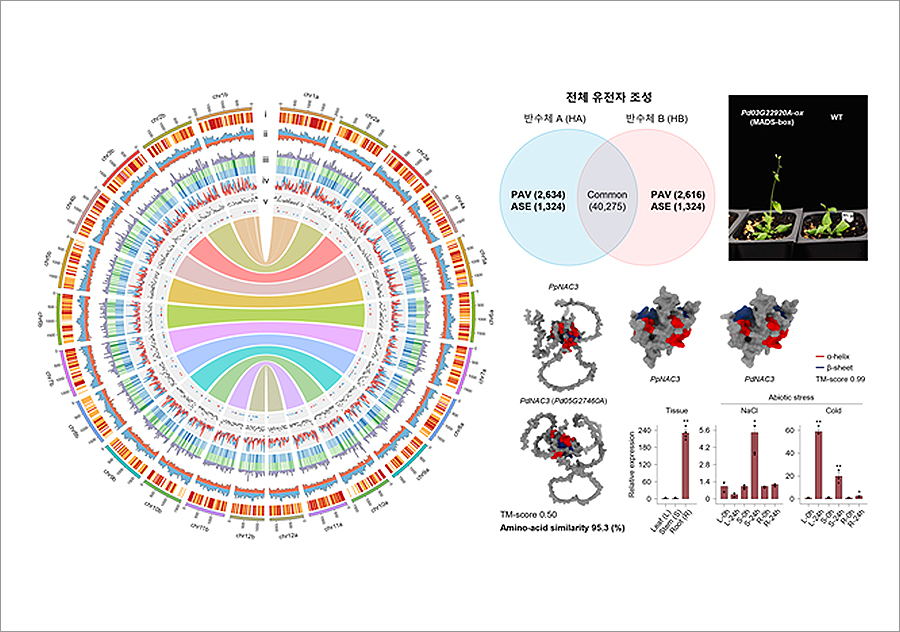
▲ Part of the Study on the Use of Genetic Information of Pine Tree Jungipumsong
The findings were published online in Nature Genetics (IF=31.7), the most prestigious international journal in the field of life sciences genetics on October 20, 2024, titled in ‘Haplotype-resolved genome assembly and resequencing analysis provide insights into genome evolution and allelic imbalance in Pinus densiflora’

▲ The picture of pine tree Jungipumsong
The study is the first of its kind among the outer plants to build a standard genome of a pine tree's haploid unit and attempt to explore genetic variation of the haploid-based wild individuals. The research team analyzed the dielectric variation of pine trees in detail based on the haploid genotype information and identified genetic differences that occur during the evolution of the giant dielectric.
Most plants exist as diploids consisting of two pairs of haploids, one from the mother and the other from the father. Previously, the difference between the two haploids was not considered to be significant, so previous studies were conducted by integrating them into one dielectric information, but this study analyzes the two haploids respectively based on the fact that the genetic difference between the haploids is very large.

▲ University of Seoul research team. Professor Kim Seung-il (left), Dr. Jang Min-jeong (middle), and Dr. Cho Hye-jeong (right)
The research team constructed two sets of high-quality haploid genomes totaling 21.7Gb (more than seven times the human genome) using Phasing techniques, the latest genome assembly method, targeting Korea's iconic pine tree "Pinus dentiflora" (Jungipumsong). It is evaluated as the highest quality among the outer plant genomes released so far and contributed to identifying the evolutionary mechanisms of genome rearrangement and important functional transcription factors in the process of species differentiation in pine trees.
In addition, as part of a study on genetic diversity in pine trees, the research team attempted to analyze sequence variation that reflected haploid information on 30 wild species of pine trees in Korea for the first time. Through this, they revealed the effect of individual genetic variation on pine gene diversity, suggesting that data construction reflecting the dielectric information of multiple individuals is necessary.
Professor Kim Seung-il said, "This study has significant meanings on the aspect of laying an important foundation for breeding on pine trees and other major trees. Also it is expected to accelerate follow-up research in various fields such as pine nematode resistance, pine seed breeding, and carbon reduction."
The study was carried out with the support of the Korea Research Foundation's Emerging Research Support Project, the Ministry of Agriculture, Food and Rural Affairs' Digital Breeding Conversion Technology Development Project, and the Forest Science Technology Development Project of the Korea Forest Service.







